.png)
Classification and Regression Trees
| Short Description | Decision Tree Regression and Classification, as a machine learning technique, is commonly used in urban health research to predict health-related outcomes or understand the importance of health-related variables |
|---|---|
| Data | A collection of variables related to urban health |
| Suggested tools | PythonRStata |
| Category | Spatial Modelling |
| Variable | Multivariable |
Overview
Decision Tree Regression and Decision Tree Classification are machine learning algorithms commonly used in urban health research to analyze and predict various health-related outcomes or variables. Decision Tree Regression models the relationship between various urban factors (like air quality or access to green spaces) and a continuous health outcome, aiming to produce precise numerical predictions. Decision Tree Classification is used when the health outcome is categorical, such as categorizing urban areas into low, medium, or high health risk based on factors like pollution levels, healthcare access, and socioeconomic status, helping in identifying regions that require more healthcare resources or environmental interventions.
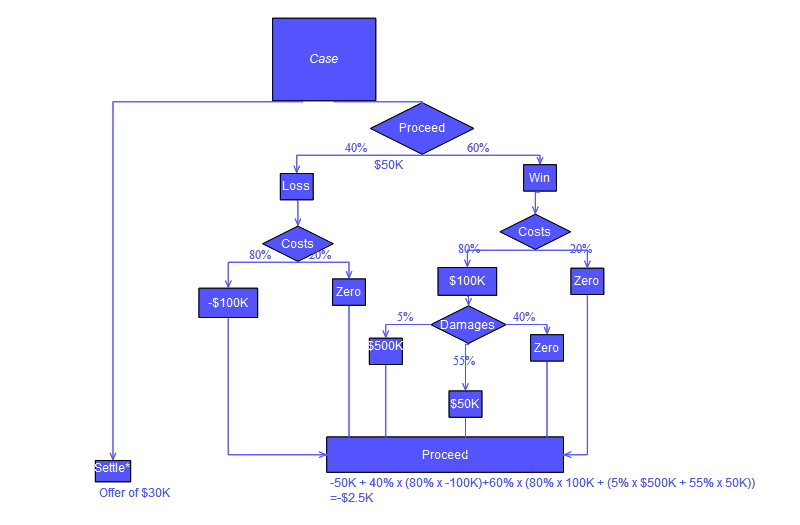
Single Decision Tree: A simple model that splits the data based on feature values to minimize variance within each node. It's interpretable but can easily overfit the training data.
Random Forest Regressor: An ensemble method that uses multiple decision trees to reduce overfitting and improve prediction accuracy by averaging the predictions of individual trees.
Gradient Boosting Regressor: Sequentially adds new trees that correct errors made by previously trained trees to minimize prediction error, often leading to higher accuracy than Random Forests but at the cost of being more prone to overfitting.
Extreme Gradient Boosting (XGBoost) Regressor: An optimized gradient boosting library that is more efficient and effective than the traditional Gradient Boosting method. It includes regularization parameters to control overfitting.
LightGBM: A gradient boosting framework that uses tree-based learning algorithms designed for speed and efficiency. It splits the tree leaf-wise rather than level-wise.
CatBoost: A gradient boosting algorithm that handles categorical variables very well and reduces the need for extensive data preprocessing.
Description
- Data Preparation: Collect and preprocess the dataset, ensuring that it contains the relevant predictor variables and the target variable (e.g., a health outcome measure).
- Building the Tree: Construct the decision tree by recursively partitioning the data based on predictor variables. The goal is to minimize the variability within each resulting partition.
- Splitting Criteria: Choose a splitting criterion, such as mean squared error or variance reduction, to determine the optimal variable and value for partitioning the data.
- Terminal Nodes: Define a stopping criterion, such as a minimum number of data points per terminal node or a maximum depth of the tree.
- Prediction: For new data points, traverse the decision tree and predict the target variable by averaging the values of the training instances in the corresponding terminal node.
Tutorial
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeRegressor, DecisionTreeClassifier
from sklearn.metrics import mean_squared_error, accuracy_score
import pandas as pd
# Load your dataset
# This is a placeholder step; replace it with loading your actual urban health dataset
data = pd.read_csv('path_to_your_dataset.csv')
# Define your features and target variable
# Let's assume 'health_outcome' is our target; for regression, it's continuous; for classification, it's categorical
features = data.drop('health_outcome', axis=1)
target = data['health_outcome']
# Split the dataset into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(features, target, test_size=0.2, random_state=42)
# For Regression
regressor = DecisionTreeRegressor(random_state=42)
regressor.fit(X_train, y_train)
y_pred_reg = regressor.predict(X_test)
reg_mse = mean_squared_error(y_test, y_pred_reg)
print(f'Regression MSE: {reg_mse}')
# For Classification
# Ensure your target variable is categorical for this part
classifier = DecisionTreeClassifier(random_state=42)
classifier.fit(X_train, y_train)
y_pred_class = classifier.predict(X_test)
class_accuracy = accuracy_score(y_test, y_pred_class)
print(f'Classification Accuracy: {class_accuracy}')
library(rpart)
# Assuming you have a dataset 'urban_health_data.csv' with various urban health-related variables
# and a target outcome 'health_outcome'. For regression, 'health_outcome' is continuous;
# for classification, it's categorical (e.g., 'Low', 'Medium', 'High').
# Load your dataset
data <- read.csv("path_to_your_urban_health_data.csv")
# Split your data into features and target
features <- data[, -which(names(data) == "health_outcome")] # exclude the target variable
target <- data$health_outcome
# Split data into training and test sets
set.seed(42) # for reproducibility
train_indices <- sample(1:nrow(data), size = 0.8*nrow(data))
train_data <- data[train_indices, ]
test_data <- data[-train_indices, ]
### For Regression
# Fit a regression tree
fit_reg <- rpart(health_outcome ~ ., data = train_data, method = "anova")
# Predict on test set
predictions_reg <- predict(fit_reg, newdata = test_data)
# Evaluate the model (e.g., using Mean Squared Error)
mse_reg <- mean((test_data$health_outcome - predictions_reg)^2)
cat("Regression MSE:", mse_reg, "\n")
### For Classification
# Fit a classification tree (assuming 'health_outcome' is now a factor for classification)
train_data$health_outcome <- as.factor(train_data$health_outcome) # Ensure the target is a factor
fit_class <- rpart(health_outcome ~ ., data = train_data, method = "class")
# Predict on test set
predictions_class <- predict(fit_class, newdata = test_data, type = "class")
# Evaluate the model (e.g., using Accuracy)
accuracy_class <- sum(predictions_class == test_data$health_outcome) / nrow(test_data)
cat("Classification Accuracy:", accuracy_class, "\n")